The Viral Most Wanted: The Orthomyxoviruses
The Spanish Flu—the Great Influenza pandemic of 1918-1919—caused by a virus called influenza type A subtype H1N1. It and its many life-threatening relatives are part of the Orthomyxovirus family.
- 12 June 2024
- 11 min read
- by CEPI
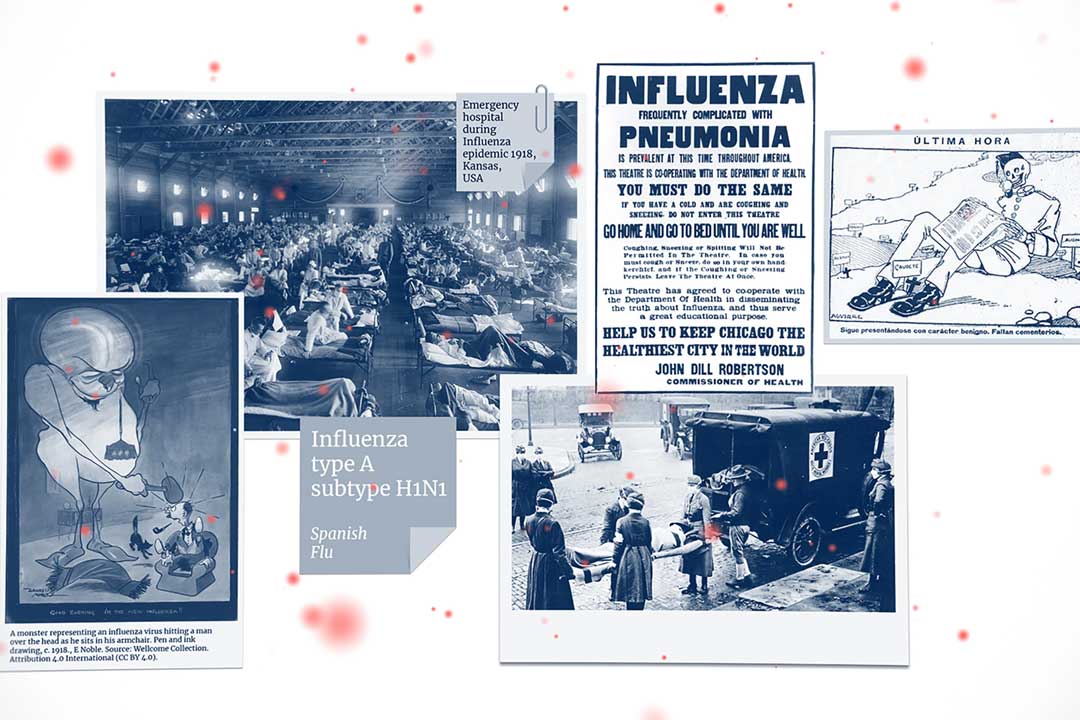
It's considered the most deadly single contagious disease event in all of human history and infected 500 million people worldwide—a third of the planet's population.
It struck in three merciless waves—the first, relatively mild, in March 1918; the second, more brutal, in August 1918; and the third and deadliest of all in late 1918, running through the first few months of 1919.
It is estimated to have killed almost five times as many people as were killed during World War One.
More than half of the people it killed were fit, strong, healthy young adults in the prime of life—including millions of World War One soldiers.
Its victims often died within hours or days of developing symptoms. Their skin would turn blue and their lungs would fill with fluid, causing them to suffocate from within.
This was the Spanish Flu—the Great Influenza pandemic of 1918-1919—caused by a virus called influenza type A subtype H1N1. It and its many life-threatening relatives are part of the Orthomyxovirus family—one of The Viral Most Wanted.
One Big Close-Knit Family?
The Orthomyxoviruses certainly make a very large family indeed. It consists of seven subgroups: the Alphainfluenzaviruses, the Betainfluenzaviruses, the Gammainfluenzaviruses, the Deltainfluenzaviruses, the Isaviruses, the Thogotoviruses and the Quaranjaviruses. The first four of these subgroups contain many virulent influenza viruses that can cause severe and sometimes deadly respiratory infections in many types of animals and birds.
Prime Suspects
By far the most fearsome members of the Orthomyxovirus family in terms of threats to humankind are those in the Alpha-, Beta-, Gamma- and Deltainfluenzavirus subgroups. These are the subgroups of viruses behind outbreaks of seasonal influenza in birds and mammals, including people, as well as the multiple deadly flu pandemics that have hit humanity over the last century.
Among the many scores of influenza viruses, several strains are already notorious for their potential to cause epidemics and pandemics. The most alarming of these are the influenza A viruses, followed by the influenza B viruses. Only influenza type A viruses are known to have caused pandemics. The lethal strain of H1N1 influenza that caused the deadliest-ever pandemic in 1918 and 1919 is thought to have killed up to 500 million people. A cousin H1N1 strain then caused another large but significantly less deadly pandemic almost 100 years later in 2009. A strain called h3N2 caused a pandemic in 1957 and was also the prime suspect behind a previous flu epidemic in 1889. Another pandemic flu strain is H3N2, which caused a milder pandemic in 1968. Disease detectives are also watching the H5N1, H7N9 and H7N7 strains because of their dangerous potential to spillover from birds and/ or animals and cause infections in people.
Nicknames and Aliases
The first and most common alias used within the Orthomyxovirus family is "flu"—short for influenza.
The H1N1 flu strain that caused the 1918 pandemic often goes by the nickname Spanish Flu and is sometimes also known as The Great Influenza. It was also called "Purple Death" because of its ability to turn its victims' skin blue and purple as their lungs filled with fluid and they suffocated. In other nations and cultures, this prolific killer was known under other aliases. In America, people talked of "three-day fever". In France it was known as "purulent bronchitis", while in Italy it was "sand fly fever." In Germany, hospitals filled with victims of Blitzkatarrh or "Flanders fever".
The related H1N1 strain behind the 2009 pandemic was originally nicknamed Mexican Flu and subsequently often referred to as Swine Flu. h3N2 has the alias Asian Flu, while H3N2 is also known as Hong Kong Flu.
The Hs and Ns that feature in the names of these Alphainfluenza viruses, or influenza A viruses, stand for the two glycoproteins contained within each virus particle—Hemagglutinin and Neuraminidase—and signify certain infectious characteristics of the viral strain.
Distinguishing Features
Orthomyxovirus virions, or virus particles, are generally between 80 and 120 nanometres in diameter and around 300 nanometres long. They are 'enveloped'—meaning they are cloaked in a membrane. The Orthomyxovirus virion envelope has small clusters and spikes of two key glycoproteins—Hemagglutinin and Neuraminidase—sticking out of it, which helps it attach to cells. These glycoproteins give the individual Orthomyxoviruses their H- and N-based nicknames.
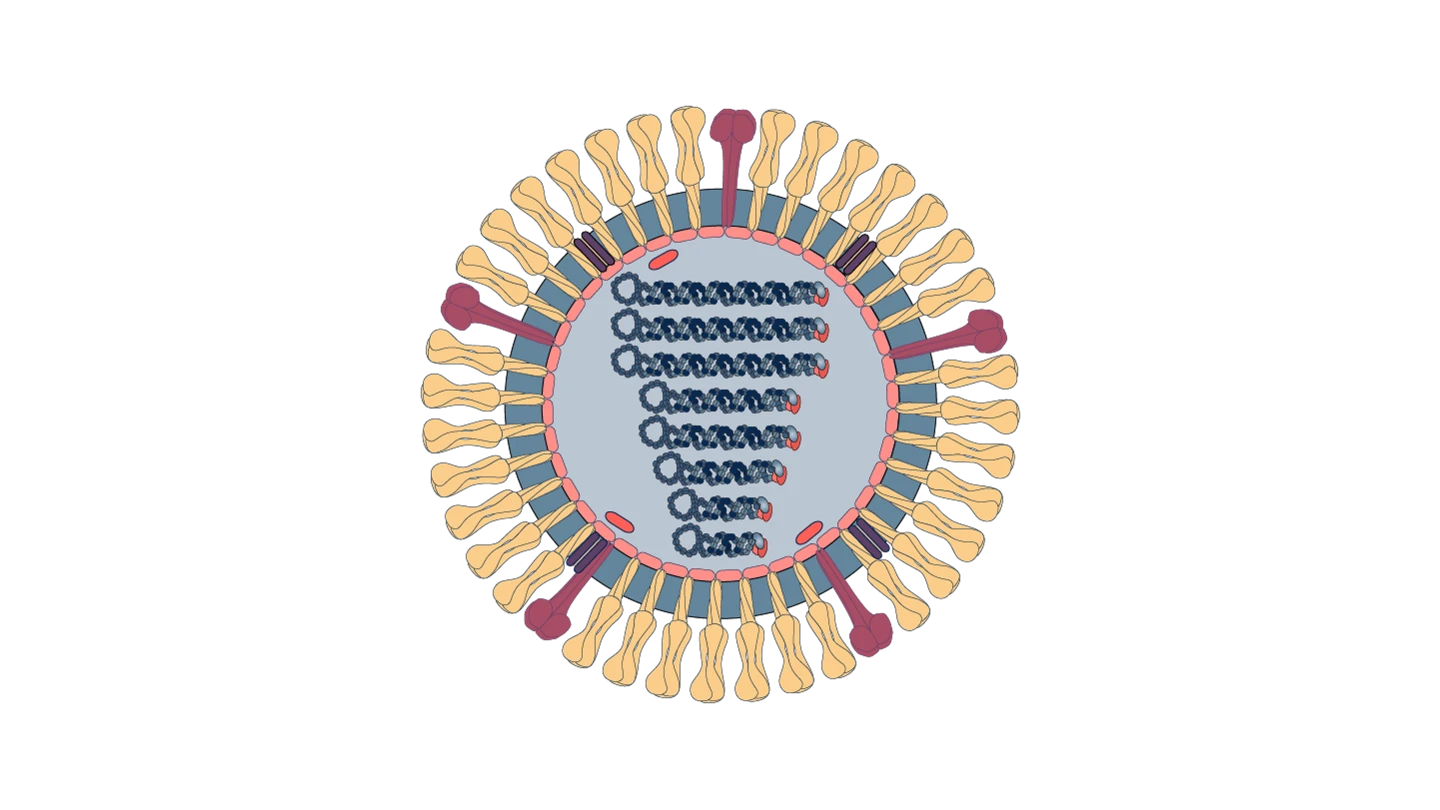
Source: Viral Zone by SwissBioPics
Modus Operandi
Orthomyxoviruses, including the notorious and deadly influenza viruses, use the H Hemagglutinin glycoprotein spike to attach to the surface of cells. They specifically target cells called epithelial cells that form a shield-like lining in the throat and the lungs. Once the virus is attached to a cell, the cell engulfs it and pulls it inside. There, the virus then sheds its cloak or envelope, hijacks the host cell's machinery, and begins to replicate.
Accomplices
Members of the Orthomyxovirus family have accomplices everywhere. From domestic pigs to ticks and mosquitoes, and from farmed poultry to wild birds such as ducks and swans, these fast-spreading viral attackers have plenty of partners in crime. Some accomplices of influenza can carry viruses thousands of miles as they migrate to new regions of the world. And because they can be infected with and carry some flu strains without getting sick themselves, they act as silent spreaders.
The influenza viruses that infect people—including seasonal strains and pandemic strains—also use human behaviour to spread. Many influenza viruses are highly contagious and are transmitted in the tiny droplets of fluid that come out when people cough and sneeze. These tiny droplets containing infectious viral particles can travel as far as a metre through the air and contaminate frequently touched surfaces such as door handles, taps or mobile phones.
Some non-influenza Orthomyxoviruses—such as Thogoto Virus and Dhori Virus—use insects like ticks and sometimes mosquitoes as vectors to help them spread to other animals, including people.
Common Victims
Influenza viruses from the Orthomyxovirus family can affect a very wide range of people, but certain groups are more likely to fall victim to an attack that makes them severely ill, or even kills.
Because their immune systems are still developing or because their immune systems are weakened by age, babies and young children under five years old and adults over 65 years or older are flu's most common victims, particularly in poorer countries where health systems are less robust. Data from the Global Burden of Disease study show that 99 percent of influenza-related deaths in children under five years old with influenza-related lower respiratory tract infections are in developing countries.
Weaker or compromised immune systems also make pregnant women and people with chronic health conditions more likely victims of Orthomyxovirus influenza infection. Some pandemic influenza strains—such as the 1918 Spanish Flu for example—buck the normal trend of seasonal flu outbreaks and hit strong healthy young adults hard.

Infamous Outbreaks
By far the most infamous of all influenza outbreaks—and arguably of all infectious disease outbreaks in recent history—was the Spanish Flu pandemic of 1918 to 1919. Infecting a third of the world's population and killing between 50 and 100 million people, this influenza A H1N1 Orthomyxovirus was a merciless and relentless killer that caused a truly global catastrophe. With no vaccines to protect against any kind of influenza and no antibiotics to treat secondary bacterial infections, efforts to control this pandemic were limited to non-pharmaceutical interventions such as social distancing and isolation, quarantine, personal hygiene, use of disinfectants and bans on or advice against large public gatherings.
The next influenza A virus to hit the world and cause a deadly pandemic was h3N2. Emerging in East Asia, it was first reported in Singapore in February 1957, then in Hong Kong in April 1957. Like so many other Orthomyxovirus influenzas, it spread like wildfire and was being detected in coastal cities in the United States by the summer of 1957. Disease investigators estimate the global human death toll from this flu pandemic at 1.1 million—meaning it was significantly less deadly than its 1918 predecessor.
Ten years on, another influenza A assassin emerged in the same region of Asia and was named Hong Kong Flu. Scientists believe the H3N2 influenza A Orthomyxovirus that caused this outbreak was a close cousin of the h3N2 virus that had hit in 1957, and there is some evidence that populations who were previously exposed to the Asian Flu 10 years earlier had some immunity to the Hong Kong pandemic strain. Ultimately, this infamous outbreak turned out to be one of the least deadly pandemics of the 20th Century, although the death toll was an estimated 750,000 to one million.
Have you read?
The most modern of flu pandemics came in 2009 when the influenza A H1N1 virus returned and, this time, became known as Swine Flu. The first identified human case was in the town of La Gloria in Mexico. Analysis by disease detectives showed that this Orthomyxovirus family member was a new strain of H1N1 that was a triple reassortment of bird, swine and human flu viruses. The strain then further combined with a Eurasian pig flu virus—which was what gave it its new nickname. The virus spread rapidly across the world and caused hundreds of millions of cases of infection in people of all age groups. Because in most cases the disease was relatively mild or even asymptomatic, the 2009 H1N1 pandemic became infamous more for the way governments and populations responded to it than for the number of people it killed. In that response, several international pharmaceutical companies developed new vaccines that could reduce the severity of disease. But because the vaccines were slow to manufacture at scale, and because many wealthy countries had pre-agreed deals to buy up large quantities of doses for their own populations, the vast majority of lower and middle-income countries had little or no access to them.
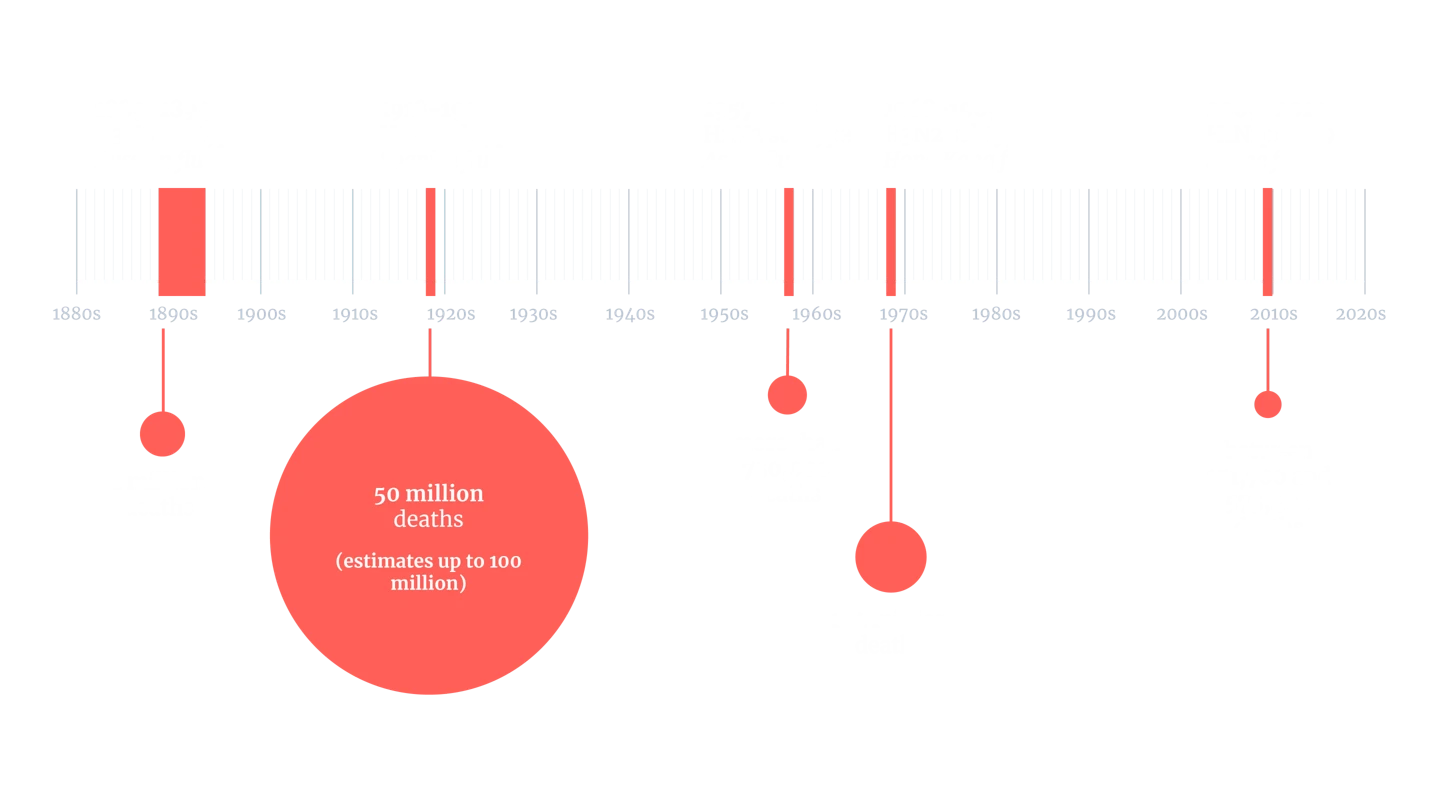
Note: The global mortality rates displayed are approximate estimates, based on the best available data and generally accepted figures at this point in time.
Common Harms
The most threatening human Orthomyxoviruses are the influenza viruses—including both seasonal flu and pandemic flu strains—that target the respiratory system and can cause severe, sometimes fatal, disease. The most common initial harms are symptoms of high fever with chills and sweats, muscle aches, intense headache, eye pain, cough, sore throat, shortness of breath, fatigue and weakness, congestion and—more commonly in children than adults—vomiting and diarrhoea.
In severe cases of infection with particularly vicious viruses—such as the H1N1 Spanish Flu for example—the disease took hold so rapidly that some victims could go from being healthy and well in the morning to dead before the end of the day. Within hours of experiencing initial symptoms of fever and headache, many victims developed pneumonia and cyanosis—when the skin turns blue due to a shortage of oxygen—before haemorrhages also caused catastrophic vomiting and nosebleeds. Ultimately, victims died of suffocation from fluid flooding their lungs.
Lines of Enquiry
Since scientific understanding of infectious diseases at the time of the 1918 Spanish Flu pandemic was relatively limited, initial lines of enquiry into how to combat or protect against Orthomyxovirus influenza attacks went down the wrong path by pursuing a bacterial rather than a viral suspect.
It was only in 1933, when the influenza virus was identified by British researchers at London's National Institute for Medical Research, that any real progress could be made towards developing a vaccine against flu. Around a decade later, the first flu vaccines were developed by disease defence experts in the United States.
These first flu vaccines themselves opened up new lines of enquiry when, in the 1947 flu season, U.S. researchers discovered they were ineffective against the flu viruses circulating at the time. This led disease detectives to the discovery that the many different types of influenza viruses that already existed and would continue to emerge would need to be targeted either with different vaccines, or one that included several flu strains.
Now, seasonal flu vaccines are manufactured each year and offered in many wealthy nations to try and minimise the impact of winter flu. But these vaccines need to go through 'strain switch' in order to be suitable for use against influenza viruses with pandemic potential because those pandemic viruses will be novel strains – a time consuming process that takes at least 4-6 months.
Scientists pursuing pandemic prevention have developed several vaccines against influenza viruses that could cause a pandemic—including strains like the influenza A H5N1 bird flu strain that has in recent years spread from birds to many types of mammal, including cattle in the United States. While at least four of these so-called pandemic preparedness vaccines are authorised for use, they take a long time to manufacture in large amounts. As a result, disease experts are now focussing their efforts on creating these and other pandemic preparedness flu vaccines using rapid-response technology such as the mRNA technology used in some of the successful COVID-19 pandemic vaccines. The vaccine makers Moderna and Pfizer both have experimental influenza A (H5N1) vaccines developed using mRNA technology in late-stage human trials.
In response to news of isolated cases of people becoming infected during an H5N1 bird flu outbreak in cattle in the United States in 2024, CEPI's pandemic watchers initiated several steps that could accelerate equitable access to H5N1 vaccines, if it were to begin to spread more swiftly in people and pose a pandemic threat. These included starting work on manufacturing preparedness and designing antigens against H5N1 using state of the art methods pioneered during the COVID-19 pandemic.
The ultimate goal of much scientific enquiry into Orthomyxovirus influenza strains is the development of a universal flu vaccine—one that could protect against many or all currently known subtypes of Orthomyxovirus influenza virus, and potentially against future novel emerging ones too. Experts in the United States had encouraging results in 2022 in initial tests of an experimental mRNA vaccine that showed broad protection against several lethal flu strains.

Written by
Kate Kelland
Website
This article was originally published by CEPI.