Uncovering the Exposome: An Emerging Field Casts a Wide Net
Researchers have long advocated for a more systematic approach to environmental health, but funding may not hold.
- 16 June 2025
- 14 min read
- by Undark
The lab buildings of Long Island’s renowned Cold Spring Harbor Laboratory have hosted researchers responsible for some of the most consequential scientific leaps in human genetics and disease. In the last 60 years, eight of the lab’s scientists have earned Nobel Prizes, including for work on how viruses replicate, on chromosome structure, and how edited genes can cause disease.
That setting felt auspicious when nearly two-dozen people, most of them researchers, gathered at a conference center about 5 miles from the main campus in December 2023 to lay out the definition for a budding scientific discipline nearly two decades in the making. The work aims to understand human health outcomes by cataloguing the tens of thousands of chemicals and conditions a person is exposed to over the course of their life and determine how those interactions affect their unique biology, a concept referred to as the exposome.
Exposome researchers say the work could be as consequential for human health as the decade-plus-long effort to read and map the human genome, or even more so, if it can realize its promise. Some experts, though, caution that even with the most recent technical advances, collecting meaningful data and sorting out how it all connects remains a significant — and expensive — challenge.
The specific contribution of genetic factors on the risk of developing many human diseases can vary widely.
And most conditions are attributable to a complex soup of genetic, environmental, social, and chemical factors. Rather than the traditional environmental health approach of studying a single type of exposure at a time, exposomics takes an intentionally wide — and exceedingly complex — view. The exposome, some researchers say, has the potential to clarify the causes of complex diseases like Parkinson’s and various cancers, and curate something that has so far proven elusive for patients: precision medicine.
Despite the enormity of its potential implications, the idea took two decades to get off of the ground and it has gotten little public attention outside of scientific journals. Until relatively recently, the technology and techniques required to catalogue a human exposome were underdeveloped (and some experts stress that there are still real limits to such approaches). But various experts in the field say that they now have better tools for helping to identify the chemicals that crop up in blood and urine samples, as well as to help crunch the massive datasets. Those advances, along with the proliferation of more powerful computational tools like artificial intelligence, have created significant momentum in recent years.
“Our ability to achieve this big vision has become much more of a reality in the last few years than it was earlier,” said Rima Habre, an exposure scientist at the University of Southern California’s Keck School of Medicine. The evolution of the field, she said, “is very exciting because we finally have tools that can help us get there in these big data sets.”

In September, Habre and other U.S. researchers launched a collaboration, led by Columbia University and funded with more than $1.5 million in support from the National Institutes of Health, just months before the Trump administration began hatcheting funds for federal research projects. The new network will support the development of analytical tools needed for exposomics research — including measurement and modeling methods for location-based exposure and high volume chemical analysis — and help U.S. researchers work with partners around the world. The U.S. effort corresponds with international collaborations recently launched in Europe, and a growing global interest in establishing a framework for the field this summer.
Despite the federal funding cuts, many exposomics researchers remain hopeful. Funding for the U.S. center remains untouched, and many of its core researchers suggested in interviews that their goals cross political boundaries. As Gary Miller, a Columbia University toxicologist, put it: “I don’t know if apolitical is the right term for it, but it’s a very scientific and rigorous approach to studying human disease.”
Cancer epidemiologist Christopher Wild first proposed “an ‘exposome’ to match the ‘genome’” in a 2005 paper. Similar to a person’s unique genetic makeup, the exposome would describe, Wild continued, “life-course environmental exposures (including lifestyle factors), from the prenatal period onwards.”
Wild’s concept generated more of a ripple than a splash among environmental health researchers. (Even he admits that a few years after publication, the paper received “virtually no attention.”) Decades later, scientists still point to his work as the defining start to the field, but it took four years for the term to reappear in scientific literature, when a group of French researchers published a paper looking at disease across nearly 60,000 occupational health reports. A few months later, the U.S. National Academy of Sciences hosted a workshop to whip up more interest, but the field lacked the methods it needed to truly flourish.
“I don’t know if apolitical is the right term for it, but it’s a very scientific and rigorous approach to studying human disease.”
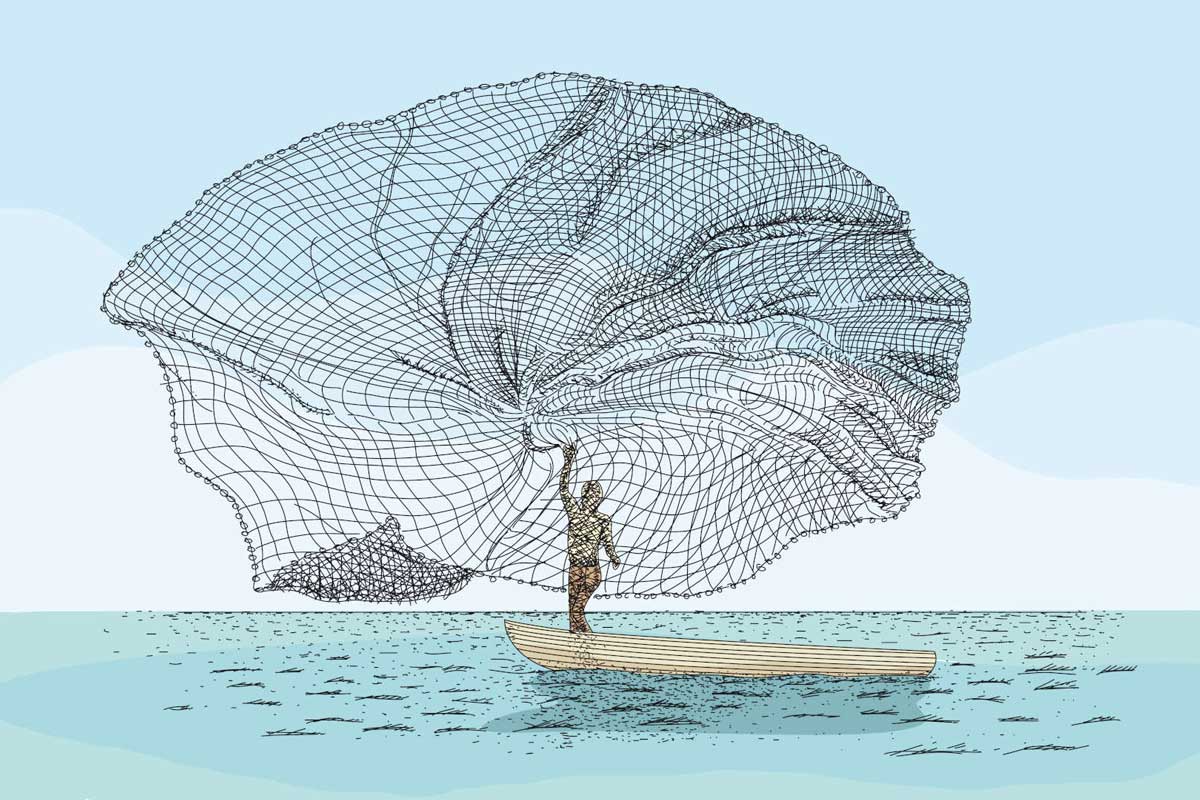
In traditional environmental health research, scientists focus on a single identifiable compound or type of exposure (lead, for example), and investigate whether it’s associated with particular health outcomes (like cancer or IQ or kidney disease). If the traditional approach is like fishing in the ocean with a single pole, according to environmental analytical chemist Lee Ferguson, exposomics is akin to hauling up nets and nets of critters. Scientists must then sift through that haul to understand its distinct components, in attempts to understand how they influence the entire ocean.
But actually building that net is very difficult, in part, because many exposures are fleeting and dynamic and there are simply so many “fish” to catch. In the earlier days of the field, “the idea of measuring all of the things you’re exposed to, it seemed impractical and impossible to do,” said Miller, the Columbia researcher, who also founded the scientific journal Exposome.
Drawing specific conclusions about the effects of exposures is an additional challenge, requiring huge sample sizes, potentially up to millions of people, said Alan Hubbard, the head of the biostatistics division at University of California – Berkeley, adding that such studies may be too unwieldy and expensive to carry out.
A few years before that 2010 National Academy of Sciences workshop, Dean Jones, a biochemist at Emory University, was at work on methods to help make such studies possible. He was studying oxidative stress, a process that can cause disease and premature aging when the body tires of fighting off cell-harming free radicals. A raft of biochemistry studies targeting specific chemicals had done little to pinpoint the main cause of that stress. Jones said he wanted to cast a much wider net, looking at many more exposures to understand how they may stress cells: “I stepped back from that targeted experimental research to ask the question, ‘Well, how can we look at this more globally?’”
At the time, tools called mass spectrometers — which identify compounds based on molecular weight — were largely used to target known compounds in human samples. Before testing a blood or urine sample, researchers usually purified them to pull out unrelated and unknown compounds. After attending a meeting that discussed the potential to run “untargeted analysis” — which looks at all contaminants in a given sample — Jones began running similar tests with unpurified samples. Rather than identifying a handful of exposures, these untargeted tests could identify tens of thousands of exposures and related compounds, including those derived from microplastics, forever chemicals, and heavy metals.
Jones said many researchers were opposed to measuring compounds that were unrecognizable. “We fought that uphill battle,” he said, making clear that “just because you didn’t know what it was, didn’t mean that it was not important.”
He and his collaborators — including Miller, then working at Emory — saw the glimmer of potential. Using those techniques, they hoped to catalogue the human exposome in the same way that the Human Genome Project had catalogued genes. In 2013, Jones and Miller helped launch Emory’s HERCULES Exposome Research Center, the first exposomics-focused center in the U.S.
Similar work was underway abroad; the European Union disbursed its first exposomics-focused grants in 2012. As in the U.S., researchers there were grappling with understanding the impact of the environment on disease, said Roel Vermeulen, an epidemiologist at Utrecht University and a leader of the International Human Exposome Network, a global effort based in Europe. In addition to employing traditional epidemiological approaches, he said, scientists saw the need for a “more agnostic way to see, can we find things that we didn’t appreciate before?”
Still, it took more than a decade for the tools needed to study exposomics to mature. In the last 15 years, Vermeulen points to what he called a revolution in wearables — portable sensors, like silicon wristbands, which track human exposures to chemicals as people move around the world — remote sensing, data capacity, and data processing. These tools made it easier for scientists to gather data on interactions with chemicals including organic pollutants, pesticides, and cosmetics, and understand where they are occurring in space and time.
The resolution of mass spectrometers sharpened as well, said Miller at Columbia, who is also a principal investigator at the new Network for Exposomics in the U.S., or NEXUS. Miller likens the advancement to when microscopes improved enough for scientists to clearly view cells.
If the traditional approach is like fishing in the ocean with a single pole, exposomics is akin to hauling up nets and nets of critters.
Researchers can now measure thousands of molecules in a small vial of human blood, said Miller, searching for potential indicators of disease — a specific compound that indicates inflammation, for instance — across wide populations. “We’re letting the mass spectrometer tell us what we should be looking at,” he said.
Such advances could help researchers study environmental exposures in specific places, like in East Palestine, Ohio. Scientists from Harvard University are currently investigating the impacts of the Norfolk Southern train derailment there, which resulted in the release of hazardous chemicals including incomplete combustion byproducts like dioxins. And the approach may also scale up to cover entire continents. In 2019, Vermeulen began a study that aims to examine the exposome data of 55 million people across Europe.
Once exposomics data is collected, making sense of it is a bit like sorting through a pile of bricks to construct a building. If it works, researchers will have genomic data and exposure data from blood and urine samples, spatial data from air or other geographic sensors, and many other types of data like a person’s proximity to a grocery store, their job, or their education level. One 2023 exposomics study, for example, examined incidence of Covid-19-related hospitalization. In addition to incorporating the electronic health records of more than 50,000 patients in Florida, the study drew from datasets on walkability, food access, and neighborhood green space to understand how environmental factors may influence the severity of the disease.


(Right) A view of the inside of a gas chromatography-mass spectrometry instrument, showing the coiled up column that samples pass through before entering the mass spectrometer. Visual: Courtesy of NEXUS

Epidemiologists and other environmental health researchers use statistical models to draw meaning from data, generally showing correlations between a single exposure, or a small group of exposures, and disease. But those calculations become much more complex as the number of variables increases. Exposome researchers have proposed several methods that may help deal with this rush of data: “Dimension reduction” picks out the variables from untargeted testing to those most likely to be linked with other data — connecting particulate pollution with asthma, for instance.
Researchers are also looking to artificial intelligence to help digest the onslaught of data. In an April report to the European Parliament on exposome research, Vermeuelen wrote that AI could analyze environmental exposures along with clinical and genetic data to predict disease risk, or pick out potential biomarkers of disease. Algorithms could also be used to track environmental data, like pollution levels, in real-time. But any use of AI requires validated and quality-controlled data, cautions Jana Klanova, an environmental chemist and coordinator for Europe’s Environmental Exposure Assessment Research Infrastructure. “If those existing resources are crap, then it is garbage in, garbage out,” she said.
Exposomics experts want to ensure the data they collect serves a purpose, but they generally favor gathering more information rather than less. Thus far, Vermeulen’s European study has scanned more than 10,000 samples for both environmental exposure and internal bodily processes. Though the study aims for an even larger cohort, he said to understand the drivers of disease at the E.U. level will require data on at least 10 million people. Most modern large cohort studies include a maximum of hundreds to thousands of people.
That massive scale is one of exposomics biggest promises, but also a huge challenge, and it has led to skepticism about the field’s practical utility. The conference held near Cold Spring Harbor was meant to tackle one of the field’s core difficulties: If exposomics encompasses every exposure, social condition, and interaction over an entire lifetime, how can researchers ever fully define and map it?
In April, Miller and the group of researchers who attended the meeting published a paper in Science that they hoped would help answer some questions about the field, including how to harmonize methodologies across labs and integrate exposomics into existing studies. The meeting, some attendees said, was a “pivotal event” and a “major milestone” for the concept’s future. But some other researchers remain concerned that the scope could hamper the field, especially given the current funding landscape in the U.S.
To grow the field, “we need a more focused approach,” said Heather Stapleton, an environmental chemist and exposure scientist at Duke who helps lead Duke’s Environmental Exposomics Laboratory. Funding traditional exposure studies is expensive as is, and Stapleton estimates that a more wide-reaching exposomics study could cost millions of dollars. Even before recent cuts to federal funding, she said, most relevant grants at the U.S. National Institutes of Health topped out at $500,000 per year for no more than five years.
The massive scale is one of exposomics biggest promises, but also a huge challenge, and it has led to skepticism about the field’s practical utility.
Plus, various estimates suggest society has engineered tens of thousands — possibly many more — chemicals, bits of which are floating all around the world and may enter the human body. Identifying them all and how they impact people is a herculean task, made even more difficult by the fact that the exposome is changing constantly as we move about the world. Integrating social elements, like childhood trauma and socioeconomic background, are another essential element of understanding the exposome, but they draw in even more information.
Have you read?
“It’s already complex if you’re just looking at a couple of exposures and maybe the impact on health through genomic mechanisms,” said Hubbard at Berkeley, who has co-authored exposomics papers but said he is still unclear on what the term means. “I think the principle is good for what we’re after, but for guiding specific scientific studies, I think it has less utility, because you can’t study everything. We don’t have enough data.”
Exposomics researchers generally view its complexity as a boon, however. The field — some researchers view it as more of a framework or paradigm — is intentionally broad and aspirational.
“We will never be able to reconstruct all exposure over the lifetime for everyone. It’s not possible and will never be possible. But we can try to compile, as much as possible, all these different factors,” Vermeulen said. He emphasized that it’s not about completeness.
Vermeulen points to the record of the Human Genome Project as an indicator of how powerful even partial exposomic data could be. When the project formally ended in 2003, scientists had sequenced an incomplete genome, but it had wide influence: demonstrating genetic similarities between humans and the model organisms often used for medical research, for example, and allowing for the sequencing of diseases and viruses. Similarly, for the exposome, “even a partial characterization would offer major opportunities to understand disease etiology,” wrote Wild in an April paper examining the 20-year anniversary of the term.
In May, Vermeulen helped gather hundreds of attendees in Washington, D.C. for an “Exposome Moonshot Forum.” Part of the program focused on developing working groups and roadmaps that could work towards a Human Exposome Project.
Those involved in the field hope to share methods and standards across borders. Klanova and Vermeulen are among the European researchers working to increase collaboration across continental lines, in part to ensure data sharing and uniform methodology. Launched the year before the U.S. effort, the International Human Exposome Network hopes to have an interim roadmap in place later this year to guide international strategy on future exposome research, which includes working with the new NIH-funded center.
“We all see that we need to go this way, just given the simple fact that environmental drivers, in the broadest sense of the word, are really the largest driver of the burden of disease.”
At this point, it’s unclear if that joint effort will be threatened by funding cuts in the U.S. Many researchers in the field say there is general uncertainty about what may happen next, though, so far, only a few have had lost grant money. David Balshaw, director of the Divison of Extramural Research and Training at the National Institute of Environmental Health Sciences, said the office is in regular communication with counterparts in Europe. But in response to a request for comment on the funding status of the NEXUS center, Emily Hilliard, press secretary at the Department of Health and Human Services, said that the NIH is unable to comment on the status of specific grants due to ongoing litigation, in an email she asked be attributed to an NIH spokesperson. (For one key NIH leader, there may be a personal connection to the work: Agency director Jay Bhattacharya served as a graduate school mentor to Chirag Patel, now a Harvard researcher leading exposome initiatives in the U.S.).
“I think we of course do have a globally different political landscape,” said Vermeulen. “But I think we all see that we need to go this way, just given the simple fact that environmental drivers, in the broadest sense of the word, are really the largest driver of the burden of disease.”