Microbes with memory: why some infections fight back
Bacteria may carry a ‘memory’ of past environments that they pass down through generations, creating subpopulations that resist treatment.
- 27 August 2025
- 3 min read
- by Linda Geddes
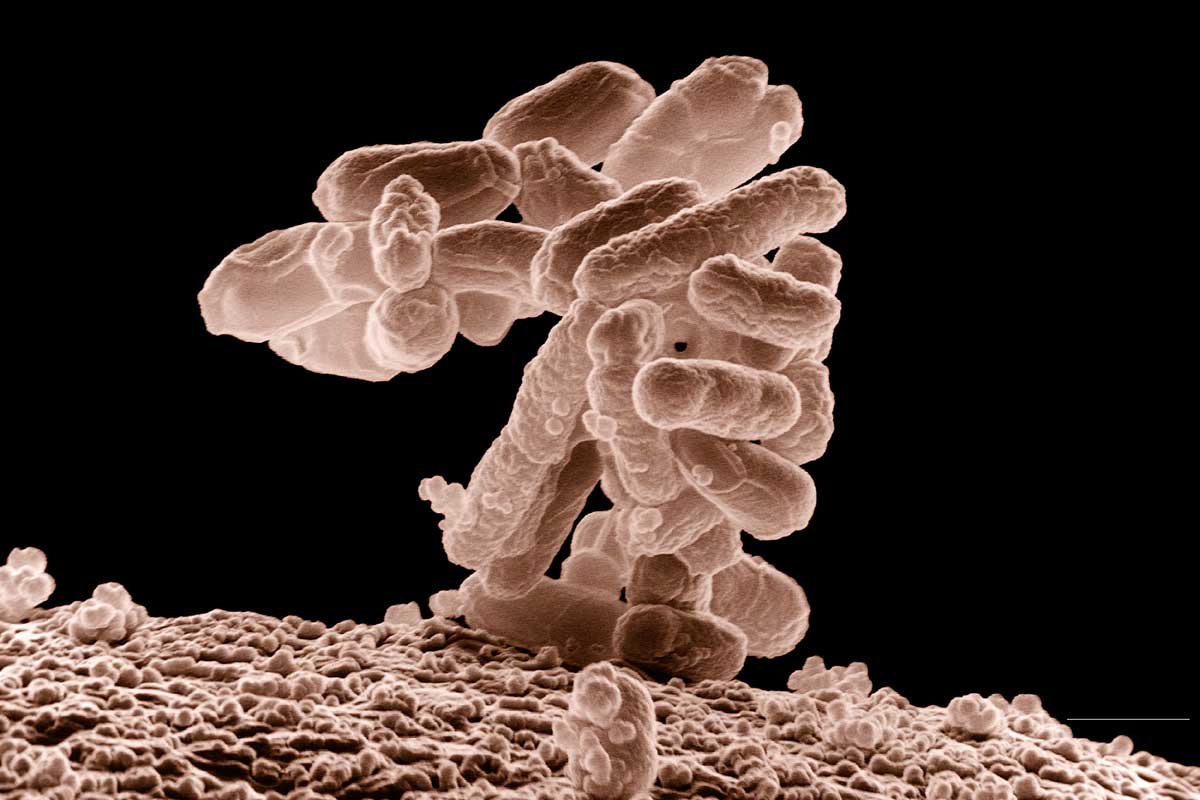
Like families passing down traditions, bacteria can pass down ‘memories’ of past environments, it seems. A single cell can hand this memory to up to 20 generations of offspring, creating subgroups of bacteria within infections that behave differently from one another. These findings, from a recently published study, could help explain why antibiotics and vaccines sometimes fail – and may point the way toward more precise treatments in the future.
“We’ve been treating bacteria as if they’re all the same, but in reality, even a single cell carries a story of its past,” said Dr Raya Faigenbaum-Romm at the Hebrew University of Jerusalem in Israel, who led the research.
Identically different
For years, scientists have puzzled over a curious quirk of bacteria: even when they share the same DNA, they don’t always act alike. Some multiply quickly, others lag behind. Some are seemingly impervious to antibiotics, while their neighbours perish.
Previously, people had assumed that a population of bacteria was largely identical, with perhaps a few rogue mutants carrying antibiotic resistance genes. There had been clues that the situation might be more complex, with genetically identical bacteria behaving differently – perhaps in response to the specific environment they find themselves in.
There were also signs that these differences might occasionally be passed to future generations, e.g. through epigenetics – a process where tiny chemical ‘tags’ attach to DNA and change how genes are turned on or off, without altering the DNA code itself. However, other differences were thought to be just random ‘noise’ that quickly disappeared.
The tricky part has been figuring out which changes last, and which don’t. Techniques like single-cell RNA sequencing can capture a snapshot of gene activity in individual bacteria, but destroy the cells in the process, making it difficult to track whether a particular state endures.
To get around this, Prof Nathalie Balaban at the Hebrew University of Jerusalem in Israel and colleagues have developed a technique called Microcolony-seq. By letting single bacteria grow into tiny colonies, and running genetic tests on all of them, they can see which changes are inherited across generations and which ones fade away.
Bacterial memory
Using this method, they discovered that, even within a single infection, disease-causing bacteria like Escherichia coli and Staphylococcus aureus split into stable subpopulations that pass on certain traits. Because these traits probably reflect how the bacteria responded to earlier conditions, the researchers describe them as a kind of bacterial ‘memory’.
“What we found is that even a single bacterium carries a long-lasting memory of where it’s been. When it divides, its descendants preserve that memory – sometimes for 20 generations or more,” said Faigenbaum-Romm.
This means that infections thought to be made up of identical bacteria could be mixtures of different groups, each with their own role. For instance, in urinary tract or bloodstream infections, some offspring were observed to have activated mechanisms that helped them cling to host cells, while others switched on genes that enabled them to move about or survive in harsh conditions.
The research, published in Cell, also suggests that this microbial memory may have limits: once nutrients in their environment became depleted, the bacterial populations apparently reset themselves.
Have you read?
Clinical implications
The researchers believe their findings could have clinical implications. Conventional clinical tests that sample just one colony could easily miss these hidden players, leading to the failure of treatments, such as antibiotics. This may help explain why so many experimental drugs and vaccines against S. aureus infections have stumbled in clinical trials: they targeted only one part of the bacterial population, leaving others untouched.
“An infection is rarely a uniform population of bacteria. It’s more like a coalition of different players, each with its own strengths," Balaban said. "To design therapies that truly work, we need to understand – and target – all of them.”